The Streichan lab studies morphogenesis, the process by which developing multicellular organisms obtain their shape. We combine cutting-edge microscopy and quantitative, physics-inspired analysis and modeling to study how cells dynamically coordinate to generate forces and shape tissues.
The discipline of physics has had striking success in uncovering quantitative laws governing the inanimate world - just think of celestial mechanics. A comparable understanding of the living world, combining quantitative observation and predictive, mathematical models, is still lacking. Our ambition is to uncover the quantitative laws that govern animal development, holding ourselves to the standards of physics. And just as a solid knowledge of electrodynamics and statics is key for any engineer, a quantitative understanding of morphogenesis opens the door to synthetic developmental biology.
Members of our lab work on a diverse set of questions in multiple systems, based on shared technology and expertise.
Model systems
Stem-cell derived organoids
Understanding human organ formation is a major scientific challenge. We develop chip-based culture systems to study the self-organization of micropatterned stem cells, which transform into precise three-dimensional cell-fate patterns and organ shapes. Recently, we used this approach to recreate neural tube folding from human stem cells in a dish. This method of reproducible organs-on-a-dish enables a quantitative approach to identify underlying mechanisms and probe the response of these systems to geometric perturbations. Check out our recent paper on this topic here. Lab members: Eyal Karzbrun, Aimal Khankhel, Matt Lefebvre, and Markus Merk.
Drosophila melanogaster
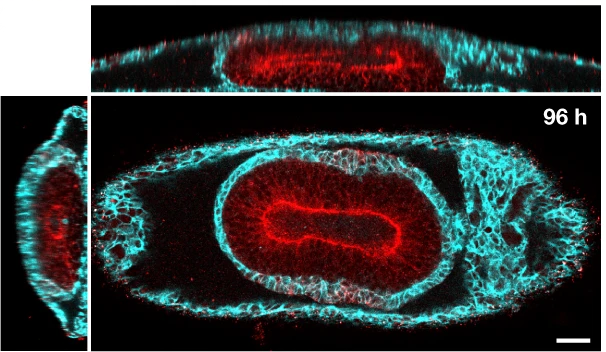
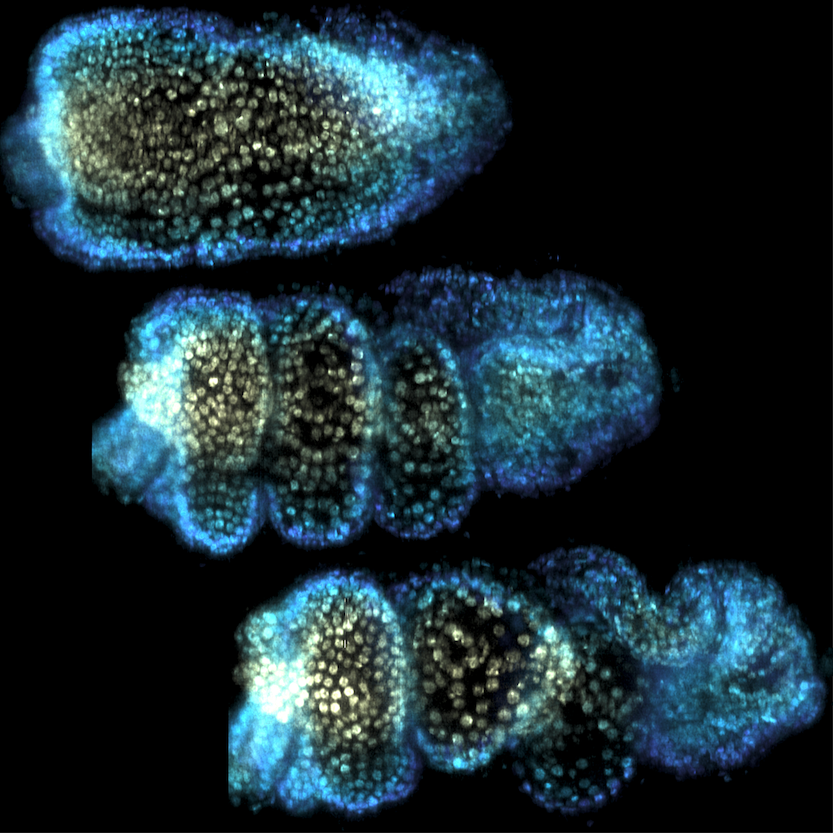
A major effort in our group addresses the physical processes underlying the early movements of the fly embryo. During gastrulation, fly embryos separate themselves into the three tissue layers of endoderm, mesoderm, and ectoderm. The ectoderm elongates by a factor via a process called convergent extension which occurs in a great number of developmental contexts, for example in the neural plate. Our lab studies how the motor molecules driving convegrent extension are controlled. We uncovered evidence that motor molecules are organized by mechanical feedback loops and geometric cues. Check out the preprints here and here. Lab members: Matt Lefebvre, Hannah Gustafson, Noah Mitchell, and Nikolas Claussen.

We are also developing a dynamic atlas of Drosophila morphogenesis which digitally stitches multiple live and fixed recordings into a single 'virtual embryo' for which information on the spatial pattern and temporal dynamics of many key genes can be queries. Lab members: Noah Mitchell and Vishank Jain-Sharma.
We also have pioneered quantitative study of the fly gut, a tube that folds into a coil of compartments. By tracing the dynamics of this organ, we identified a mechanism of 3D shape change relying on a mechanical program across tissue layers. Gene expression patterns govern the emergence of calcium pulses in the muscle layer, triggering shape change of the adjacent endoderm layer to fold in 3D. Check out the preprint here. Lab members: Noah Mitchell and Yuzheng Lin.
Zebrafish
We have recently added the zebrafish Danio Rerio to our model systems. Zebrafish are transparent, making them a perfect model to observe vertebrate development. We aim to image live zebrafish in toto using lightsheet microscopy. Lab members: Susie Wompat.
Parhyale
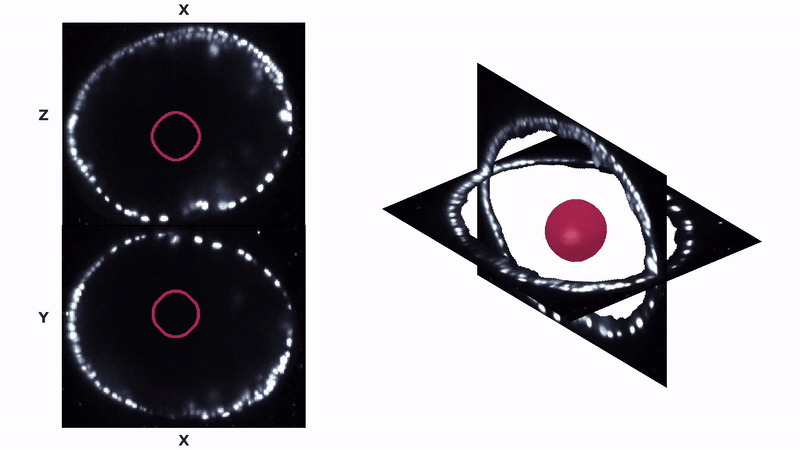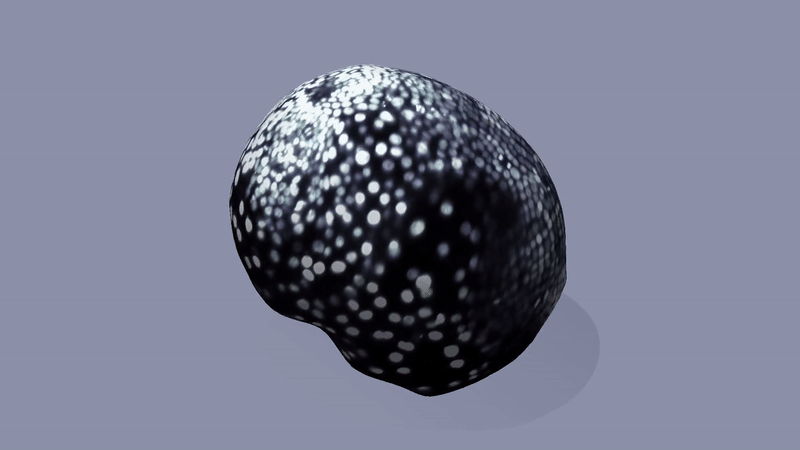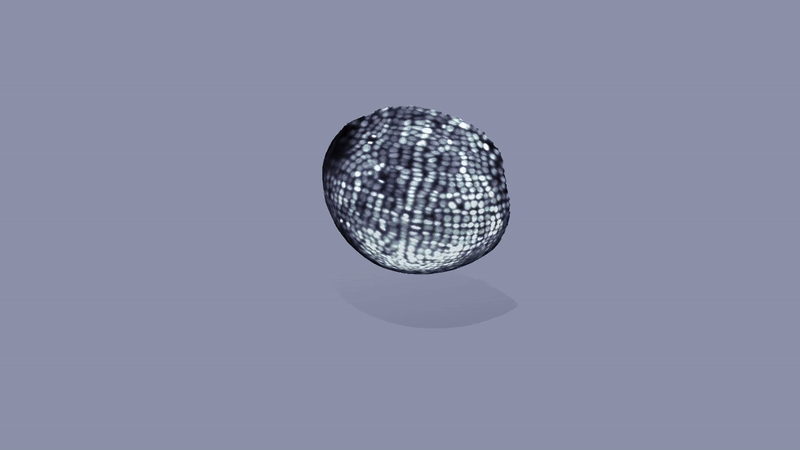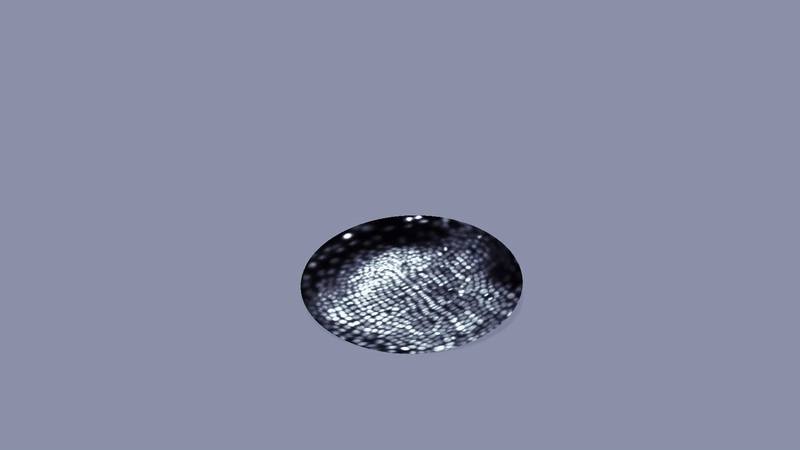
Parhyale hawaiensis is a small crustacean and an emerging model for limb morphogenesis. Our research has elucidated how active, directed cell division creates an usual, 4-fold order in this organism. Check out the preprint here. Lab members: Dillon Cislo.
Hydra
Hydra are a class of simple freshwater organisms which are remarkable for their capacity of regeneration: a hydra cut into piece can regenerate its entire body plan. How this happens remains incompletely understood, with tantalizing evidence for a key role of geometry, topology, and mechanical feedback. Lab members: Fridtjof Brauns and Eyal Karzbrun.
Methods
Microscopy & Computational Imaging
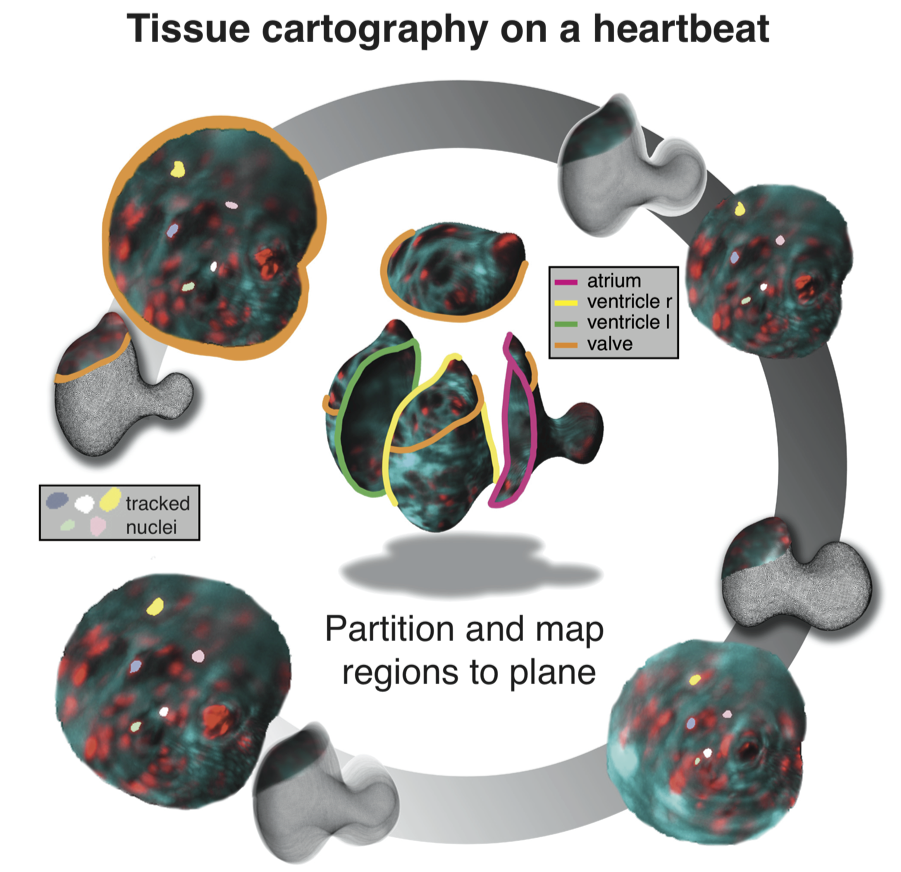
To understand morphogenesis, it is key to have access to dynamic data showing the entire embryo, and to be able to carry out sophisticated quantitative analysis. Our group has pioneered methods for rapid in-toto imaging of embryos and organs using light-sheet microscopy and methods for cartographic analysis of 3D morphogenetic processes.
We are currently in the process of constructing a so-called light field microscope capable of imaging entire sample volumes at once and achieve in toto imaging with sub-second resolution. Lab members: Pieter Derksen.
Quantitative modeling
In collaboration with Boris Shraiman (Kavli Institute for Theoretical Physics), we develop mathematical, predictive models of morphogenesis based on physics-inspired ideas. We have shown that this type of "theoretical biology" can make interesting and successful predictions: dynamic recruitment, a form of mechanical feedback in which cells recruit motor molecules in response to deformation, was first predicted based on theoretical considerations (see paper) before being demonstrated experimentally (preprint). We believe that mathematical models are a key to enable quantitative and rigorous testing of biological hypotheses.
Optogenetics
We recently used optogenetic perturbations to demonstrate mechanical feedback during embryonic development in the fly. By shining light on a tissue, we induce mechanical stress and study the behavior of the motor molecule myosin in adjacent cells, which are subjected to strong forces from the optogenetic activiation. Check out the preprint here.
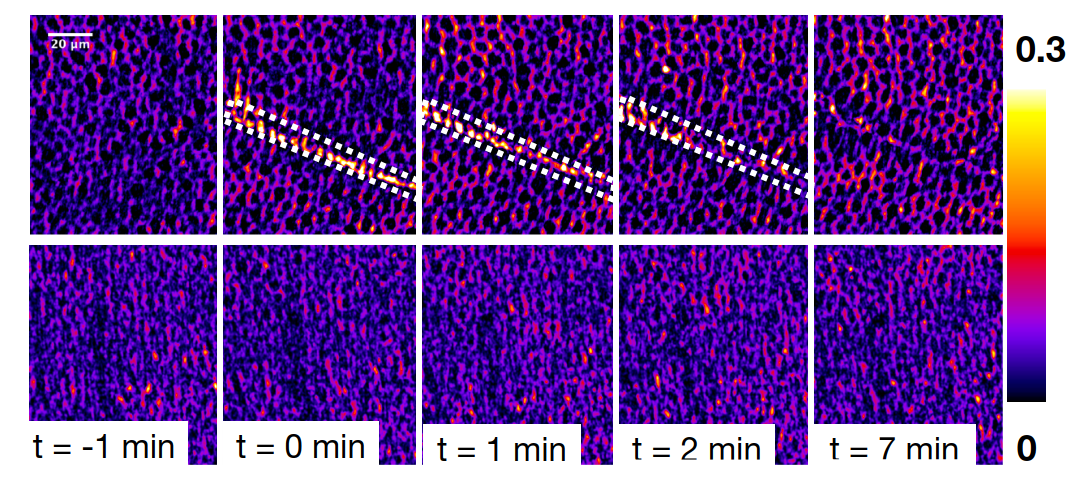
We used similar optogenetic tools to study mechanical action of tethered tissue layers during organogenesis. Check out the preprint here.
Organs on a dish
In our work on organoids, we have pioneered methods to fold synthetic organoids in a reproducible way by micro-patterning the dishes on which they grow. Check out the protocol here.